by
Josť Aguilar-Manjarrez
This appendix provides full and detailed descriptions of the database and the methodology used in this study. Compared with the first fish farming GIS study done for Africa, the methodology developed here is considerably more sophisticated. This study has been based on the development of a recent study for Latin America. In comparison to the latter, the present one has made adjustments and improvements. The most significant advance has been the development of the fish growth models.
INTERNET addresses are provided in this appendix for the interest of readers who wish to obtain the original data that were used in this study or for additional details.
Hardware
Almost the entire GIS analyses was carried out using a Sun Spark Station 10.
Other GIS analyses and ASCII data manipulations using QBASIC and VISUAL-BASIC programs were run on a Pentium PC, MS P/133, 1.2 GB, 17 inch Video Super VGA. The PC was set up to run under DOS with no network connection to obtain maximum memory (e.g. for running QBASIC programs) or with network capabilities for data file transfers.
Data transfers between the Sun Spark Station 10 and the Pentium PC were carried out under Microsoft Windows for work groups version 3.11 and XTGOLD software.
Digital data for verification of fish farming potential in Uganda were created by digitizing a paper map using a Calcomp 9100 SMART A0 digitizer.
Software
A Geographic Information System (ARC, Version 7.0.3, ESRI, Redlands, CA, USA) which has both raster and vector capabilities was used. This system is able to efficiently store geographically referenced information in a database which includes both digital maps and their attribute files. Different criteria were kept in separate directories in the database and were called when required. Criteria were combined using logical conditions and/or mathematical operations according to the models developed.
Other GIS software (i.e. IDRISI for Windows version 1, ERDAS version 1 and IDA version 4.2) were used to take advantage of some of their capabilities (e.g. MCE from IDRISI) or as a tool to convert to or from different data formats (e.g. UNIX - ERDAS - IDRISI file conversions or vice versa).
Base grid
A grid was used as a template to standardise grid extensions. Since most of the study was based on the air temperature data, the cell size and geographic projection of one of these data were chosen as the base grid. A copy of the Grid file for maximum temperature for the month of January was made, renamed as AFMASK and reclassified to contain only values of 1 for the African continent and -999 for the NO DATA values (e.g. the Ocean).
A description of AFMASK is illustrated below:
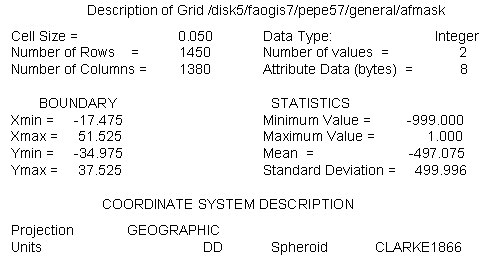
Arc: describe afmask
The spatial extensions of AFMASK ensure coverage of the entire African continent except for Madagascar. Data originally stored in vector or ASCII format or having a different resolution or projection were converted to the base grid.
Based on the number of rows and columns defined above, the base grid had a total of 2,001,000 pixels (i.e. 1450 * 1380 = 2,001,000). The number of pixels for the African continent including the inland water bodies was 1,004,351 (i.e. Madagascar not included).
Database directory
The present database was almost entirely developed under the directory: /sun2disk5/faogis7/pepe57/ of SUN2. Due to lack of space in the latter directory, part of the appendix was developed under the following directories: /sun1disk3/faogis3/waba/ and /sun1disk3/faogis3/epsanex of SUN1.

As with any GIS-base study, the analysis was strongly dependent upon the quality and accuracy of the data used, so in order to account for this, a detailed description of the data used is provided below:
CONSTRAINTS
Protected areas
Delineation of protected areas in digital format (vector) were available from the WCMC. Those areas were classifies as: conservation; wildlife and additional forest. These areas were classified into a single class with a value of zero.
INTERNET address: http://unep.unep.org/unep/partners/global/wcmc/home.htm
Water bodies
They were derived from the DCW (ESRI, 1992a) at 1 million scale and rasterized. The grid includes perennial lakes and rivers which can be detected at 0.050 degree resolution. These areas were classified into a single class with a value of zero.
INTERNET address: DCW can be downloaded from the Digital Chart of the World data Server of the Pennsylvania State University Libraries. Also see the following site for DCW in ARC/INFO format: http://www.esri.com/base/data/online/browse.html
Major cities
Locations of major cities were provided by ArcWorld at 1:3000,000 scale, rasterized, and classified into a single class with a value of zero.
INTERNET address: http://www.esri.com/base/support/faq/pcarc/5datacon/7_1547.html The DCW site (above) also contains information for Arc/World.
FARMING SYSTEM MODELS
FACTORS ( PHYSICAL AND ENVIRONMENTAL RESOURCES )
Water requirement
Precipitation
The source data were twelve ASCII files containing gridded values of mean monthly precipitation. The source data and the procedure to import these files into GRID is similar to the one described for the air temperature data (below). The standard errors of the rainfall grids range between 5 and 15%, depending on the data density and the spatial variability of the actual mean monthly rainfall. These files were obtained in standard ARC/INFO ASCII INTEGER GRID format in millimetres.
INTERNET address: http://cres.anu.edu.au/software/africatxt.html
Potential evapotranspiration
Twelve GRID files containing monthly potential evapotranspiration values for the whole world in ARC/INFO export format were obtained from IIASA (i.e. IIASA-LUC project and W.Cramer).
The PET data were calculated using the Penman-Monteith equation (Monteith, 1965; 1981; Doorenbos et al., 1992) based on the IIASA-LUC project input data. In more detail, the input data were derived from two sources:
a) Leemans and Cramer (1991), and
b) Variable Wind Speed: derived from LUC project based on ECMWF database (over 10 years daily wind data) were averaged to monthly data.
INTERNET address: http://www.iiasa.ac.at/
Soil and terrain suitability for fish ponds
Soils
Soils data for Africa were obtained from the DSMW CD-ROM (Version 3.5, FAO, 1995). This database is based on the FAO-UNESCO Soil Map of the World, original scale 1:5,000,000. The CD-ROM contains two types of files: DSMW map sheets and Derived Soil Properties files with images derived from the Soil Map of the World.
The DSMW consists of the following map sheets: Africa, North America, Central America, Europe, Central and Northeast Asia, Far East, Southeast Asia, and Oceania. The maps are available in three formats: one vector format (ARC/INFO Export), and two raster formats called ERDAS and IDRISI (or flat raster) formats. All the maps are in geographic projection, with spherical datum. The coordinates are expressed in decimal degrees. The scale of the original map (and the vector formatted data) is 1:500, 000.
The Derived Soil Properties files consist of interpretation programmes and related data files. The programs are written in QuickBasic version 4.5 and can be read using a DOS or OS/2 operating system. Included are programmes that interpret the maps in terms of agronomic and environmental parameters such as: pH, organic carbon content, C/N ratio, clay mineralogy, soil depth, soil and terrain suitability for a specific crop production, soil moisture storage capacity, and soil drainage class. The output is given in the form of maps and data files which can be stored for later retrieval. Special country analyses can be made for specific soil inventories, problem soils and fertility capability classification for every country in the world. Also included are maps of classification units of the World Soil reference Base Units and topsoil distribution, which can facilitate the teaching of soil science. Finally, there is a soil database developed, specifically for environmental studies on a global scale, which includes soil moisture storage capacity, soil drainage class and effective soil depth.
INTERNET address: http://internal.fao.org/ag/agl/agls/t1.htm
Slope
Slope was derived from a DEM. A new globally consistent 1km resolution (1:1 million map scale) DEM dataset called GTOPO30 was used. This database is the product of more than three years of international collaborative efforts involving UNEP, USGS, NASA, GSI and INEGI. Data from different sources and resolutions were brought together to create this product using new algorithms and a software developed by scientists from the EROS-Data Center.
INTERNET address: http://edcwww.cr.usgs.gov/landdaac/gtopo30/gtopo30.html
LAND USES AND INFRASTRUCTURE
Livestock wastes and agricultural by-products
Livestock wastes
Livestock population was used as a surrogate for manure. Livestock populations for cattle, goats, sheep and pigs in the form of animal/km2 were obtained from U.S. Army CERL and CRSSA, Cook College, Rutgers University.
The GlobalArcTM CD-ROM Database is for use with ARC/INFO 6.0 or higher and ARC-VIEWTM 2 software. It is a global-scale environmental database, converted from all of the raster data contained in the Global GRASS CD-ROM datasets 1-5, the largest single-format, global environmental database available. This CD-ROM contains 84 themes and a total of 147 layers. CRSSA was responsible for converting the datasets to ARC/INFO format.
INTERNET address: http://deathstar.rutgers.edu/global/info.html
Agricultural by-products
Data on crops were derived from an African land cover data-set. This database is one portion of a global land cover characteristics database that is being developed at a continent-by-continent basis. All continents in the global database share the same map projection and are based on 1-km AVHRR data spanning April 1992 through March 1993.
The raster image of the Africa land cover contains class number values for each pixel that correspond to the appropriate land cover classification scheme legend. On the basis of such a classification scheme legend, a core set of derived thematic maps was produced through the aggregation of seasonal land cover regions which are included in each continental database.
The original African land cover data set contains 195 classes, from which five sets are derived: (1) Olson Global Ecosystems Legend, consisting of 94 classes; (2) IGBP Land Cover Classification, 17 classes; (3) U.S. Geological Survey Land Use/Land Cover System, 27 classes; (4) Simple Biosphere Model, 19 classes; and (5) Biosphere-Atmosphere Transfer Scheme, 20 classes.
The Africa database is available in two different map projections: the Interrupted Goode Homolosine and the Lambert Azimuthal Equal Area, the later projection was the one obtained for this study. The USGS, the University of Nebraska-Lincoln, and the European Commission's DG Joint Research Centre are generating this global land cover database, and the USGS-EROS Data Center is carrying out its distribution.
INTERNET address:
Documentation: http://edcwww.cr.usgs.gov/landdaac/glcc/globdoc.html
Data: http://edcwww.cr.usgs.gov/landdaac/glcc/glcc.html
Farm-gate sales
Population density
This layer was provided by NCGIA from the University of California at Santa Barbara. It is a grid produced by NCGIA as a result of town interpolations. The algorithm used to generate the surface also takes into account the total population of the countries and the number of people living in the 2nd level administrative districts. The GRID expresses population density in individuals/km2.
INTERNET address: http://www.geog.buffalo.edu/ncgia/W8522E00.html
Urban market size and proximity
Major cities
Locations of major cities were provided by ArcWorld (1:3,000,000). ArcWorld's major cities are represented by points, consequently even large towns do not have an area, however, since the objective of this study was to analyse the proximity to the potential market(s), the undefined extension of the towns were not a limitation as the fish market(s) could be located in any part of the town. Points are assumed to be located at the centre of towns.
INTERNET address: See major cities constraints above.
Roads
The road network was derived from ArcWorld (ESRI, 1992b) and rasterized.
INTERNET address: See major cities constraints above (roads are also included in such INTERNET site).
FISH GROWTH
Air temperature
The source data were 24 ASCII files containing gridded values of mean monthly daily minimum and maximum temperatures. Air temperature grids were created from the source, point and line data using the spatial analysis and interpolation techniques ANUDEM, ANUSPLIN and ANUCLIM developed by CRES at the Australian National University. Grids were obtained by first fitting topographically dependent climate surfaces to point climate data using procedures in the ANUSPLIN package (Hutchinson, 1991; Hutchinson and Gessler, 1994).
In addition to data already obtained by CRES from miscellaneous sources, monthly climate data were acquired from research agencies including CIMMYT, FAO, East Anglia Climate Research Unit, CSIRO Division of Forestry, Texas A & M University and from the national meteorological services of Djibouti, Gambia, Ghana, Kenya, Malawi, Morocco, Namibia, Rwanda, Seychelles, Sudan, Tanzania, Uganda and the Democratic Republic of the Congo. Data were collected over all available years of record to maximize spatial coverage. Most data were collected between 1920 and 1980, so the fitted temperature grids can be interpreted as estimates of standard means for the period of 1920 to 1980.
The number of accurately geocoded stations for which mean monthly air temperature data were obtained were 1,504 for daily minimum temperatures and 1,499 for daily maximum temperatures. Data were subjected to comprehensive error detection and corrections procedures based on ANUDEM and ANUSPLIN. Complete descriptions of these data are being prepared (Hutchinson, et al., In prep.).
The error of the temperature grids depended mainly on the accuracy of the underlying climate surfaces. The standard errors of the temperature were about 0.5 oC. Grid files were obtained in standard ARC/INFO format in units of tenths of degrees Celsius. Grids covered the entire African continent except for some off-shore islands, where climate interpolation was not supported by climate data (i.e. Madagascar). These files were processed and the output was converted to a grid (the procedure is described in Appendix 8.8).
INTERNET address: http://cres.anu.edu.au/software/africatxt.html
Wind velocity
Mean annual wind velocity was provided by UNEP/DEIA/GRID-Geneva. This file originally in IDRISI format was converted to GRID. These African data originate from manuscripts that were received by ESRI from FAO, as part of the work performed under a UNEP contract. The source maps were prepared directly from original hand-drawn maps, provided by FAO on stable basemap copied at a 1:5,000,000 scale. Isolines plotted on the FAO/AGS basemaps were then manually digitized by ESRI. The final products were rasterized at UNEP/GRID-Geneva.
INTERNET address: http://www.grid.unep.org/gnvd0039.htm
Fish growth
Predictions of fish growth were based on water temperature, in turn predicted from air temperature and wind velocity data.
INTERNET addresses:
Biosystems Analysis Group: http://biosys.bre.orst.edu/
Latin America: http://biosys.bre.orst.edu/aqua_gis/prelim.htm
Aquaculture Decision Support Software: http://biosys.bre.orst.edu/pond/pond.htm
Datum from protected areas was obtained from WCMC. The GRID file was downloaded with FTP in compressed format using UNIX 'compress' command and was called af_reg.e00.Z.
a) Uncompressing file:
gissw2-faogis>> uncompress -v af_reg.e00.Z
b) Importing the grid files:
Arc: import grid af_reg.e00.Z afreg
c) Adjusting grid extensions and cell size:
Grid: setwindow afmask
Grid: setmask afmask
Grid: setcell afmask
Grid: afreggrd = afreg1
The DCW tiles (about 100) for perennial inland water bodies for Africa were joined by K. Lethcoe of USGS. This datum was raterized at 3-arc-minutes.
Arc: gridpoly afwater afwacons_s1
Grid: citycons_s1 = merge(uncity, mscity, scity, vscity),
where: citycons_s1 = new grid with all the citycons_s1; merge = GRID command to merge GRID data; uncity = unsuitable citycons_s1; mscity = moderately suitable citycons_s1; scity = suitable citycons_s1 and vscity = very suitable citycons_s1.
Note: This data were also used in the urban market size and proximity submodel.
Re-classification of constraint grids
To mask out constraint areas from the all the maps, a single value of zero was assigned to the constraint grids and a value of one to the remaining African land area. The AML mask.aml automates these steps and combines the three grids to make an overall constraint grid:
promask1 = afreg1 (afreg1, `value gt 0')
promask1b = con(isnull (promask1) == 1, 0, promask1)
promask1c = setnull(promask1b == 1,1)
wamask1 = afwacons_s1 (afwacons_s1, `value gt 0')
wamask1b = con(isnull (wamask1) == 1, 0, wamask1)
wamask1c = setnull(wmask1b == 1,1)
citymask1 = citycons_s1 (citycons_s1, `value gt 0')
citymask1b = con(isnull (citymask1) == 1, 0, citymask1)
citymask1c = setnull(citymask1b == 1,1)
constraint = promask1c * wamask1c * citymask1c
Using this approach, when the constraints grid (constraint) is multiplied with the grids, the constraints are masked out from those grids and the suitable areas within those suitability grids retain their suitability score.
Copying and converting ASCII files into ARC/INFO GRID
Twelve ASCII files containing gridded mean monthly values of precipitation were copied and converted from the CRES compact disk into ARC/INFO grid format. Example syntax for the month of January was:
Arc: asciigrid /cdrom/961206_0935/afrain01.grd afrain01.gr
Table 8.1 shows the statistics on the monthly precipitation values in Africa and Figure 8.1 shows the monthly distribution of precipitation values.
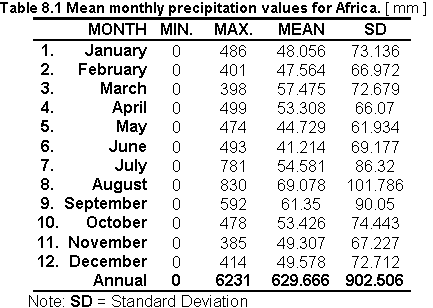
Fig. 8.1 Mean monthly precipitation
Downloading potential evapotranspiration grid files via ftp
Monthly potential evapotranspiration grid files in ARC/INFO export format were downloaded via ftp.
Preparing files for UNIX
Two steps were necessary to prepare the files into a UNIX format.
a) Renaming files: gissw2-faogis>> rename etm1.e00 etm1.e00.Z
b) Uncompressing files: gissw2-faogis>> uncompress -v etm1.e00.Z
Importing the grid files
An example syntax is illustrated below for the month of January:
Arc: import grid etm1.e00 etm1
The Grid AML gridimport.aml automates the procedures for all the months.
Adjusting grid extensions and cell size
The Grid modules SETWINDOW and SETCELL were used to extract and adjust the grid extensions of the monthly grid files to the base grid.
Grid: setwindow afmask
Grid: setmask afmask
Grid: setcell afmask
Grid: etm1gr = etm1
Moreover, because etm1 contained cells which were not a part of the temperature and precipitation grid files (e.g. Arabia) a mask had to be used to remove these areas from the evaluation.
Grid: etm1new = etm1gr * afmaskv,
where: afmask = a temperature grid file containing only one value "1".
A Grid AML etsetwindow.aml automates the procedures described above for all the months.
Table 8.2 shows the statistics on the monthly potential evapotranspiration values for Africa and Figure 8.2 shows the resulting monthly distribution of evaporation values.
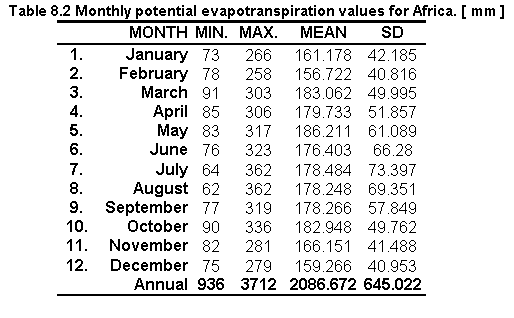
Fig. 8.2 Monthly potential evapotranspiration
a) Calculating water requirement
Table 8.3 shows the statistical water requirement values obtained by the difference between mean monthly precipitation and monthly potential evapotranspiration. The following formula was used:
WR = P - E,
where: WB = Water requirement; P = mean monthly precipitation; E = monthly potential evapotranspiration.
Example of syntax used in GIS analysis for the month of January was:
Grid: wabape = afrain01gr - etm1new
where: wabape = water derived from the difference between precipitation and evaporation; afrain01gr = mean monthly precipitation; etm1new = monthly potential evapotranspiration.

b) Calculating water requirement for shallow ponds
Based on the Latin America study (i.e. except for the evaporation coefficient) the following formula was used in order to estimate the volume of water which has to be provided to the ponds during the dry seasons:
WR = ( P * 1.1 ) - ( E * 1.3 ) - 80 ,
This formula also gives the deficit between precipitation and evaporation during that season.
where: WR = Water requirement; P= mean monthly precipitation; E = monthly potential evapotranspiration
Correction factors:
![]() precipitation was multiplied by 1.1 to
include the amount of rain that drains into the pond from the pond dikes;
precipitation was multiplied by 1.1 to
include the amount of rain that drains into the pond from the pond dikes;
![]() evaporation was multiplied by 1.3 to
compensate for the higher evaporation from free surfaces such as small open ponds;
evaporation was multiplied by 1.3 to
compensate for the higher evaporation from free surfaces such as small open ponds;
![]() 80 = seepage from ponds [ mm ].
80 = seepage from ponds [ mm ].
Example syntax of the above formula was:
Grid: waba = (afrain01gr * 1.1) - (etm1new * 1.3) - 80
where: afrain01gr = precipitation grid for the month of January; etm1= evapotranspiration grid for the month of January.
The Grid AML waba.aml automates the procedures for all the months.
The above formula was applied to the available P and E dataset and the results are shown in Table 8.4 and Figure 8.3.
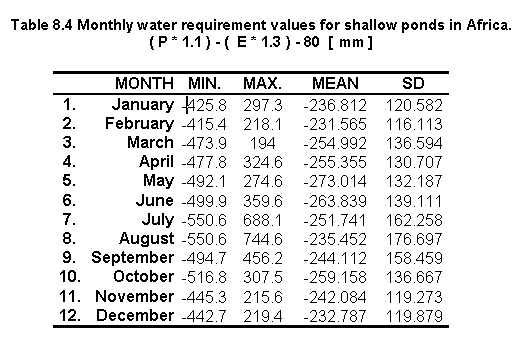
Fig. 8.3 Monthly water requirement for shallow ponds
Seasonal negative minima (creating sample points)
To evaluate the existence of two negative minima ( i.e. or areas in which the lowest minima occurs) a set of sample points was defined. These points were chosen to be at about 5o x 5 o distance (i.e. latitude) and they were used to sample the 12 monthly [ ( P * 1.1 ) - ( E * 1.3 ) - 80 ] values.
a) The Grid SAMPLE module was used to create an ASCII file listing each cell location along with the 12 monthly water requirement values.
The following syntax's was used:
Grid: Waba.txt = sample( waba1, waba2, waba3, waba4, waba5, waba6, waba7, waba8, waba9, waba10, waba11, waba12)
An extract of Waba.txt is illustrated below:
First column are the mask values not recorded in the sample point extraction; MISSING correspond to NO DATA values or -999 in this study.
b) Sample points were extracted from Waba.txt using a text editor in ARC/INFO, these points were copied into a separate file and imported to a spreadsheet for analyses.
Table 8.5 shows the statistical results of the sample points and Figures 8.4, 8.5, 8.6, 8.7, 8.8, 8.9 illustrate some examples of their distribution. It was found that seasonal minima occurred four times: latitude 0o N and S, longitude 10o (Figures 8.6 and 8.7); latitude 5 o S longitude 15 o (Figure 8.8) and latitude 10 o S, longitude 20 o (Figure 8.9).
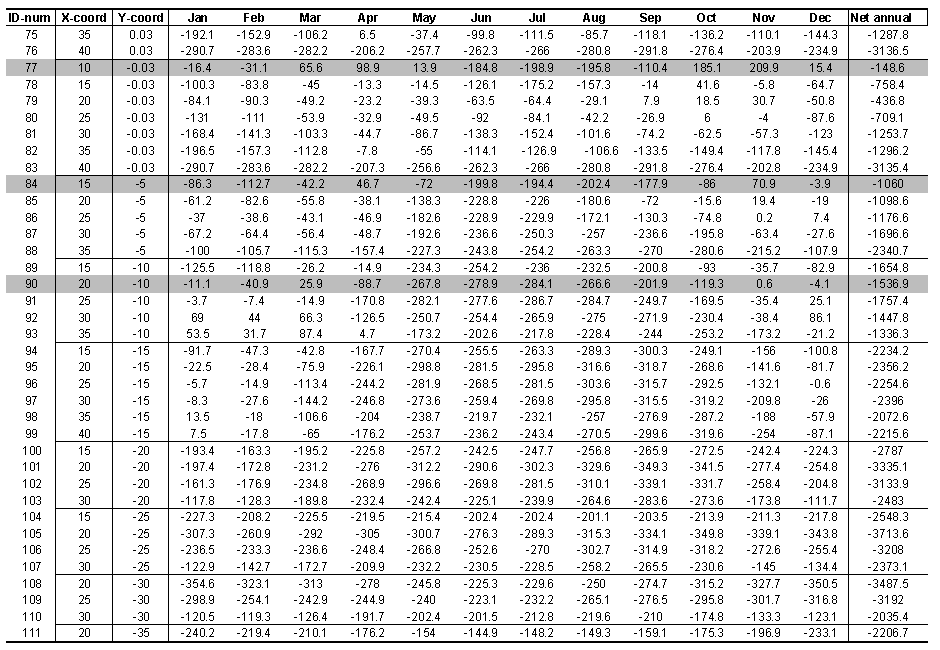
Net annual water requirement for shallow ponds
Even though two seasonal negative minima occurred four times, the total annual amount of water, which needs to be provided to the ponds during the dry seasons was found by the sum of the monthly values of [ ( P * 1.1 ) - ( E * 1.3 ) - 80 ] :
Adding the water requirement grid files:
Grid: netann = waba1 + waba2 + waba3 + waba4 + waba5 + waba6 + waba7 + waba8 + waba9 + waba10 + waba11 + waba12
where: netann = sum of the monthly water requirement grids (positive and negative values); waba1 to waba12 = water requirement grids.
Table 8.5 Water requirement sample points.
[(P*1.1) - (E*1.3) - 80] (in three parts)


Soil parameter selection, thresholds and data extraction
The appropriate soil parameter selection as well as the thresholds evaluation and data
extraction, were carried out by F. Nachtergaele (FAO-AGLS Division).
Five maps were produced: Four referred to the percentage of area covered by soils according to their suitability for fish ponds (i.e. very suitable, suitable, moderately suitable and unsuitable). The fifth map resulted in the combination of these four maps to arrive at an overall soil suitability.
Geographic projection
The maps prepared by F. Nachtergaele were in IDRISI format in plane projection. The
following document file was provided for these maps:
To convert from a plane reference system to a Lat/Long projection the following changes had to be made to the document file provided:
Exporting to ERDAS
Using the IDRISI export command, IDRISI files were converted into ERDAS format.
Converting files from ERDAS to GRID
When importing the ERDAS files into ARC/INFO using the imagegrid command, it was found
that the new geographic projection was not maintained. To solve this problem, the ERDAS
software was used. Using ERDAS, the ERDAS files were imported as IMAGE files, and then
exported as ARC/INFO GRID files.
Defining the study area (SETMASK), its extent (SETWINDOW) and cell size (SETCELL)
Figures 8.10 shows the resulting spatial distribution of the very suitable soils grid.
Converting the GTOPO30 imagine file into GRID
The DEM (GTOPO30) was obtained in IDRISI format in Lat/Long projection, so it was possible
to export it with IDRISI software as an ERDAS file and then the GRID imagegrid command was
used to convert such ERDAS file into a GRID.
Grid: imagegrid dem.gis demgrd
where: dem = ERDAS image file; demgrd = grid file.
Defining the study area (SETMASK), its extent (SETWINDOW) and cell size (SETCELL)
Creating the slopes
a) Projecting data to an equal area projection
The DEM was projected into an equal area projection (Polar quartic projection), so that
the ground units and zunits would be the same, which are in meters. No z_factor was used
for this method. The SLOPES command and the use of the zunits and the z_factor are
explained in the ARC/INFO manual.
Arc: project grid demgrd demgrd_pq,
where: demgrd = grid file; demgrd_pq = grid in an equal area projection.
b) Using the SLOPES command.
To create the slopes from the DEM the SLOPES command was used:
Grid: afslope = slopes (demgrd_pq, percentrise),
where: pndlsope = slope image; slopes = slopes command; demgrd_pq = grid in an equal area
projection; percentrise = keyword to output the percent rise, also referred to as the
percent slope.
c) Assigning thresholds
The slope grid (aflsope) was re-classified according to the thresholds defined by Hajek and Booyd (1990):
Grid: pndslope = reclass( afslope, remap_afslope),
where: pndlsope = re-classified slope grid with 4 suitability thresholds for fish ponds; reclass = grid command; remap_afslope = look up table to reclassify afslope.
Figure 8.11 shows the spatial distribution of the slope grid after re-classification.
Creating the soils and terrain suitability grid:
Grid: soilslope = 1.5 * soils5 + pndlsope
where: soilslope = soil and terrain suitability grid; 1.5 = coefficient assigned to give a higher weighting to soils5; soils5 = overall soil suitability grid; pndslope = re-classified slope grid with 4 suitability thresholds for fish ponds.
Grid: soilslope_sl = slice( soilslope, table, remap_soilslope)
where: soilslope_sl = final suitability image; remap_soilsope = look up table to reclassify soilslope.
Defining the study area (SETMASK), its extent (SETWINDOW) and cell size (SETCELL)
Example for cattle:
The same procedure was applied for the other 3 livestock types.
Data conversion from animals/km2 to livestock numbers
The cell values in the original data are not the actual number of cattle, but rather the
categories (because the data were originally created using GRASS software, which is
limited to integer cell values 0-255).
Livestock data are listed as animal/km2 x 1000. For example, category #147 (located in Kenya) is 16900 cattle/km2 x 1000 or 16.9 cattle/km2. The later option was used for this study.
a) Data for cattle, sheep, goats and pigs were projected to Flat Polar Quartic projection. Example of data conversion for cattle:
Arc: project grid cattle cattle_p,
where: project = Arc projection command; cattle = original data; cattle_pq = data in flat polar quartic projection.
Conversion of projection takes place according to the parameters specified in the file llpolq.prj illustrated below:
- INPUT
- PROJECTION GEOGRAPHIC
- UNITS DD
- PARAMETERS
- OUTPUT
- PROJECTION FLAT POLAR QUARTIC
- UNITS METERS
- PARAMETERS
- 00 00 00
- end
b) Data frequency extraction
where: tables & frequency = Arc commands for data extraction; cattle_pq = cattle data in flat polar quartic; cattle_frq = file name given containing the frequency data for cattle.
c) Data manipulation in EXCEL
To calculate the area occupied by each livestock category, the number of cells (COUNT) was multiplied by the square of the cell size in kilometres (5.930642* 5.930642). Then, this calculated area was multiplied by the livestock category.
Example: For category # 147 there are: 16.9 cattle/km2 occupying 15,167 cells (count).
Therefore:
15,167 * (5.930642 * 5.930642) = 533,462 km2
16.9 (cattle/km2) * 533,462 (km2) = 9,015,500 cattle
d) Livestock numbers from EXCEL to SUN
Calculations of livestock numbers were prepared as lookup tables in EXCEL in order to reclassify the original grid data. Once the data were prepared they were copied from EXCEL to SUN through the computer network.
Original livestock data were re-classified:
Grid: lncatlle = reclass (cattle, remap_lncattle)
where: lncattle = cattle grid containing livestock numbers; reclass = GRID command; cattle = original cattle grid; remap_lncattle = lookup table to reclassify original cattle data to livestock numbers.
Calculating the amount of manure available
a) To calculate the amount of manure available from each livestock type the following
formula was used:
Amount of manure available [tons] = livestock number [1000] x livestock weight [tons] x solid manure /1,000 kg of livestock:
b) Calculating the total amount of manure available:
totmavai = macattle + masheep + magoats + mapigs,
where: totmavai = total manure available; macattle = manure available from cattle; masheep = manure available from sheep; magoats = manure available from pigs.
c) Reclassifying into four suitability classes:
The total amount of manure available grid contained 49 values which were subdivided into 4
suitability classes.
Grid: pndmanure = reclass (totmavai , remap_totmavai),
where: pndmanure = re-classified grid image; totmavai = total manure available; remap_totmavai =
lookup table used to reclassify totmavai.
The resulting image using this reclassification is presented in Figure 8.12.
a) Converting the land cover image file into grid
The land cover image (lndcvr.img) was obtained in IDA format in Lambert Azimuthal Equal Area projection at 1 km resolution. Using a PC, the IDA version 4.2 software was used to convert the land cover image in IDA format to a GRID file using the procedure described below:
Using IDA select from the menu:
The output file, lndcvr.txt was a space delimited ASCII file containing, in each line, the pixel values of one row of the original image. The number of lines was, therefore, equal to the number of rows of the image. To convert this file to a GRID, the text file, lndcvr.txt had to be converted from DOS (i.e. PC) to UNIX format, then a header was added at the beginning of the file:
The new file, lndcvru.txt was converted to a GRID file using the ASCIIGRID command: Arc: asciigrid lndcvru.txt lndcvr, where: lndcvr = grid file.
b) Change of projection
Arc: project grid lndcvr lndcvrll lamll.prj
where: project = Arc command; lndcvr = grid file in Lambert Azimuthal Equal Area projection; lndcvrll = grid file in Lat./long projection; lamll.prj = file containing projection parameters.
Conversion of projection took place according to the file lamll.prj:
- INPUT
- PROJECTION LAMBERT_AZIMUTH
- UNITS METERS
- PARAMETERS
- OUTPUT
- PROJECTION GEOGRAPHIC
- UNITS DD
- PARAMETERS
- 00 00 00
- end
c) Defining the study area (SETMASK), its extent (SETWINDOW) and cell size (SETCELL)
d) Extracting the crops data
Efforts were made to re-classify cropland areas by crop types, but it was found that this is not possible because the data source does not differentiate cropland types. As a result, from the 195 classes provided by the original data, all cropland classes were re-classified into a single value of 4 and considered as potential sources of fish feed inputs. All other areas were assigned a value of zero (i.e. no crops): Grid: crops = reclass (acrops, remap_lndcvr), Figure 8.13 shows the spatial crop distribution.
e) Combining manure and crops
Defining the study area (SETMASK), its extent (SETWINDOW) and cell size (SETCELL)
Re-classification
Grid: farmgate = reclass (popaf_dens, remap_popden),
where: farmgate = final suitability image based on the re-classification of population density data.
Creating a grid for each of the 4 major cities
Major cities were re-classified according to their accompanying population density classification system. An example syntax is given for cities classified as very suitable:
Grid: uncity = reclass( cities, remap_uncity),
where: uncity = unsuitable cities; cities = grid containing all cities; remap_uncity = lookup table used
to reclassify unsuitable cities.
The same operation was repeated for the other 3 cities.
Creating the cost grid
The cost grid represents the time cost and it is assumed that the time required to travel from one cell to another in absence of main roads is 5 times longer than the time needed on the main road. To create such a grid the following procedures were used:
a) Roads were re-classified according to their accompanying road type classification system. An
example syntax is provided for roads classified as very suitable:
Grid: roads1 = reclass(roads, remap_roads1),
where: roads1 = very suitable roads (i.e. lowest cost) ; roads = grid containing all road types; remap_roads1 = look up table to re-classify very suitable roads.
The same operation was repeated for the other 3 road types. Areas where there was an absence of roads were assigned a value of 5.
b) Grids for the 4 roads types and a grid that represents the absence of roads were combined to create a COST grid:
rdsgrd_cost = merge (roads1, roads2, roads3, roads4, roads5),
where: roads1 = very suitable roads; roads2 = suitable roads; roads3 = moderately suitable roads; roads4 = unsuitable roads; roads5 = absence of roads.
Figure 8.14 shows the resulting cost grid:
Using the COSTDISTANCE command
This GRID command calculates the least accumulative cost distance from each cell to the
towns. In other words, it finds for each cell the best (and most convenient) route to go
to the town. It is not necessarily the closest town, but it is the easiest to reach. The
cost-distance analysis is explained in the ARC/INFO manual.
dist1_temp = costdistance(uncity, rdsgrd_cost, backl1),
where: dist1_temp = distance to unsuitable cities; rdsgrd_cost = cost grid and backl1 = grid containing the direction of the paths on a cell by cell basis.
This grid allows reconstruction of the various paths to the towns. Directions are indicated with numbers from 0 to 8 indicating the next neighbouring cell along the least accumulative cost path from any cell of the grids to reach the town. If the path is to pass into the right neighbour, the cell will be assigned the value 1, 2 for the lower right diagonal cell and so on, continuing clockwise. The value 0 is reserved for the cells representing the towns.
Changing the values of the grid backl1 into the corresponding degrees (the
North is indicated by 360 (which is equivalent to 0) )
dire1_deg = con(backl1 == 1, 90, backl1 == 2, 135, backl1 == 3, 180, backl1 == 4, 225,
backl1 == 5, 270, backl1 == 6, 315, backl1 == 7, 360, backl1 == 8, 45, backl1)
Creating a grid containing latitude values
The following GRID AML, automates the creation of the latitude grid for Africa:
where: afmask is the based grid; aflatgrd is the resulting latitude grid.
Calculating the real distances from the towns along the paths
Since the grids are in geographic co-ordinates (Latitude/Longitude in decimal degrees),
the dist1_temp grid cannot be used to calculate the distances.
Real distances can be derived from geographic distances considering that the distance between 2 points varies with the cosine of the latitude, if the direction is East-West and it is constant if the direction is North-South. Diagonal paths have a length in between the two (E-W and N-S) which vary with the sin of the direction. The formula is the following:
length1_rd1 = 111 * abs(sqrt(sqr(0.050) * (sqr(sin(dire1_deg / deg )) * cos(afrlatgrd div deg) + sqr(cos(dire1_deg / deg))))),
where: 111 is the length of 1 degree at the Equator expressed in km; 0.050 is the cell size in decimal degrees; deg is a variable built in GRID to convert from radians to degrees; afrlatgrd is a grid containing the latitude value of each cell.
Assigning weights to the road grid
Real distance grid (length1_rd1) is multiplied by the cost grid (rdsgrd_cost). This grid
is used to give a weight to the road grid in such a way that areas without roads have a
weight (cost) 5 times higher than the others.
length1_rd2 = length1_rd1 * rdsgrd_cost
The grid length1_rd2 provides weighted distances calculated from one cell to the
other
To obtain the least accumulative cost paths in real distances a new cost-distance has to
be re-calculated. The result must be divided by the cell size (0.050) to keep the already
calculated distances.
dist1_fin = costdistance( uncity, length1_rd2) / 0.050
Calculating distances in hours
Distances in hours (travelling time) is obtained by dividing the least accumulative cost
paths (dist1_fin) by 90 considering an average speed of 90 km/h on the road. Since the
road grid weighted as 4 areas without roads, it is assumed an average speed of 90 / 5 = 18
km/h outside the main roads (using graded roads or tracks).
dist1_hour = dist1_fin / 90
Limiting the suitable areas for commercial fish farming to 6 hours travelling time
dist1_h6 = con(dist1_hour le 6, dist1_hour, 9999)
The value 9999 was assigned to areas further away than 6 hours.
The 9999 value is extended to the whole African grid (covering unsuitable areas
previously indicated with NODATA)
dist1_h6a = con(isnull(dist1_h6) == 1, 9999, dist1_h6)
The same operations (steps 4.3 to 4.9 ) are repeated for the other 3 classes of
towns
The GRID AML mktgrd.aml automates the procedures for all the towns.
The least accumulative cost path grids calculated separately for the 4 classes of
towns are multiplied by the town weight and added together
cost_fin6 = min(dist1_h6a, dist2_h6a * 2, dist3_h6a * 3, dist4_h6a * 4)
Finally, the area indicated with 9999 is unsuitable
mktgrd = con(cost_fin6 ge 9999, 9999, cost_fin6)
Copying and converting ASCII files into ARC/INFO grid
Twenty four ASCII files containing mean monthly gridded values of daily minimum and daily
maximum temperatures were copied and converted from the CRES Compact disk into ARC/INFO
grid format. The following example illustrates how one of the files (AFMINT01) was copied
and converted into a grid:
Arc: asciigrid /cdrom/961206_0935/afmint01.grd afmint01.gr
Note: The GRID AFMINT01 corresponds to the month of January.
Dividing the GRID files by a factor of ten
Original ASCII files were obtained in units of tenths of degrees Celsius so the gridded
ASCII files had to be individually divided by a factor of ten to obtain their original
vales. The following example illustrates how one of the files was divided by ten:
Grid: afmint01ten = afmint01.grd / 10
Note: A Grid AML tentemp.aml automates the procedure for the 24 GRID files by repeating the command above for each one of the GRID files.
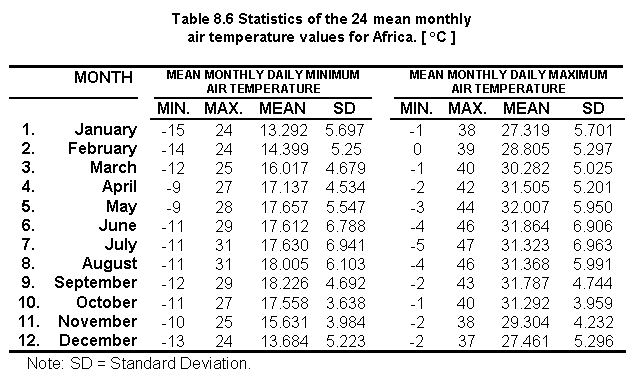
Table 8.6 shows the statistics on the 24 mean monthly air temperature values for Africa:
Merging the No Data numerical value to the GRID files
Based on the ASCII files received from CRES the No Data values in the GRID images created
were assigned a value of -999 within the African continent but the No Data values
corresponding to the Ocean (i.e. commonly called "c" values) had no numerical
value assigned to them and therefore it was necessary to assign them the No Data value of
-999 in order to create the header information necessary for the creation of the final
GRID files.
Three steps were necessary to include the -999 value in the GRID files:
a) The GRID command ISNULL was used with the base grid image AFMASK to create a GRID named TEMP. ISNULL returns a numerical value of "1" for those values that have NO DATA, and "0" if they have data, on a cell by cell basis within the analysis window. The syntax used is presented below:
Result of TEMP: African continent = 0 ; Ocean = 1
b) The GRID file TEMP was multiplied by the value of -999 to create a GRID file named NODATA.
Result of NODATA: African continent = 0 ; Ocean = -999
c) The GRID command MERGE was used to multiply the input grids (AFMINT01TEN and NODATA) into a single grid (AFMINT01NEW).
Result of NODATA: African continent = air temperatures values ; Ocean = -999
Creating two ASCII files using SAMPLE
The Grid SAMPLE module was used to create two ASCII files from the 24 GRID files above.
SAMPLE lists the values of a group of cells from one or more grids. For each selected
cell, information on cell locations in Longitude and Latitude and the relevant temperature
values from each of the input grids is written to an ASCII file.
The Grid AML gridtemp.aml was used to produce two ASCII files.
The Grid AML gridtemp is illustrated below:
MIN.TXT = sample( afmask, afmint01f, afmint02f, afmint03f, afmint04f, afmint05f, afmint06f, afmint07f, afmint08f, afmint09f, afmint10f, afmint11f, afmint12f)
MAX.TXT = sample( afmask, afmaxt01f, afmaxt02f, afmaxt03f, afmaxt04f, afmaxt05f, afmaxt06f, afmaxt07f, afmaxt08f, afmaxt09f, afmaxt10f, afmaxt11f, afmaxt12f)
Replacing MISSING with -999
A very small number of data values were reported as MISSING in both of the text files, so
these data were replaced with -999 using a text editor (i.e. find and replace option).
The ASCII files were converted into DOS format using the module UNIX2DOS in
ARC/INFO:
Arc: unix2dos min.txt afmin.dos
Arc: unix2dos max.txt afmax.dos
Creating two comma-delimited ASCII files
The QBASIC program COMMAS.BAS illustrated below was used to modify the two ASCII
files to create two comma-delimited ASCII files:
MINAF.TXT = minimum temperature data
MAXAF.TXT = maximum temperature data
The QBASIC program COMMAS.BAS is illustrated below:
CLOSE
min$ = "D:\unldatin\min.dos"
max$ = "D:\unldatin\max.dos"
minf$ = "D:\unldatin\minaf.txt"
maxf$ = "D:\unldatin\maxaf.txt"
COMMAS.BAS created two output files (MINAF.TXT and MAXAF.TXT). An extract of MINAF.TXT is reported below:
The mask values are not recorded in the output.
Calculating mean monthly air temperature
The program DATAIO.EXE was used to calculate the mean monthly air temperature. DATAIO.EXE
takes the input files AFMIN.TXT and AFMAX.TXT to create TEMPMEAN.TXT (note: file names are
user-defined; programs that can accept any name).
An extract of TEMPMEAN.TXT is reported below:
Note: TEMPMEAN.TXT was 214 Mb and took the PC 6 hours to process.
Changing the ASCII file TEMPMEAN.TXT to the format required by ARC/INFO
The QBASIC program AIRTEMP.BAS was used to change the ASCII file TEMPMEAN.TXT to the format required by the ARC command ASCIIGRID. The first part of the program checks the size of the GRID and the order of the cells. A cell out of sequence causes a program interruption and an error message. The second part checks the number of variables included in the input file and creates an equal number of output files. It also determines the number of rows and columns of the GRIDS and produces the header (see ARC/INFO ASCIIGRID syntax) indicating the items listed below:
All parameters are determined by the program itself except for the cell size and NODATA value. Default values are:
If those values need to be changed, new values must be replaced in the program (which can be edited using any available text editor) in the box indicated below:
The QBASIC program AIRTEMP.BAS is illustrated below:
Note: Each monthly water temperature files was 14 Mb. It took the PC 5 hours to create these files. An extract of one of the files AIRTMP1.TXT for the month of January is reported below:
Converting the output files of AIRTEMP.BAS to ARC/INFO GRIDS
After generating mean monthly air temperature files with TEMP.BAS, the 12 output files were converted to UNIX format (DOS2UNIX command in ARC). Once this was done, the ASCIIGRID command was used to convert the ASCII files to GRIDS. The AML airgrids.aml was used to automate the manipulation procedure for the 12 files as illustrated below:
Table 8.7 shows the statistics on the 12 mean monthly air temperature values for Africa and Figure 8.15 show the resulting monthly distribution of air temperature values.
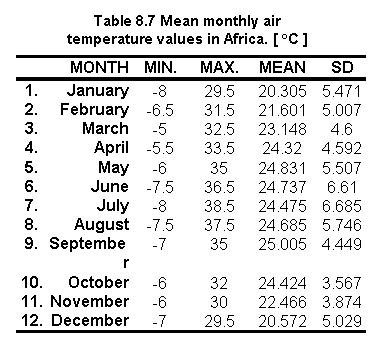
Fig. 8.15 Mean monthly air temperature values in Africa
Copying and converting the IDRISI file into ARC/INFO grid
Original data AFDBCH04.IMG was received through FTP in IDRISI format. A document
file was created for this image file in IDRISI for Windows version 1.
Using file export in IDRISI, AFDBCH04.IMG was exported as an ERDAS file as WINDAF.GIS
The IMAGEGRID module in ARC/INFO was used to convert WINDAF.GIS into a grid WINDAFGR:
Arc: imagegrid windaf.gis windafgr
Adjusting grid extensions and cell size
The Grid modules SETWINDOW and SETCELL were used to adjust the grid extensions and cell size of windafgr to the base grid:
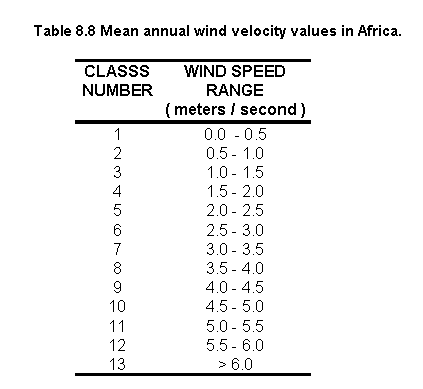
Table 8.8 shows the statistics on the mean annual wind velocity grid for Africa and Figure 8.16 illustrates the distribution.
Creating an ASCII file using SAMPLE
The following command was used to produce one ASCII file.
WINDAF.TXT = sample(afmask, windnew), where: WINDAF.TXT = ASCII file for mean annual wind velocity.
The ASCII file was converted into DOS format using the module UNIX2DOS in ARC/INFO:
Arc: unix2dos windaf.txt windaf.dos
Creating a comma-delimited ASCII file
A VISUAL-BASIC program COMWIND.BAS was created to modify the ASCII file windaf.dos
to create a comma-delimited ASCII file WINDCOM.TXT. The first part of the program
deletes the first column or mask value column. The second part places commas between the
Longitude and Latitude and the mean annual wind speed values.
WIN.BAS was executed by opening the WIND icon in program manager. A small window is open where the user can enter the input and output file locations. Example syntax:
The VISUAL-BASIC program WIND.BAS is illustrated below:
An extract of WINDCOM.TXT file is reported below:
Calculating mean monthly water temperature
The program WATEMP.EXE was used to calculate the mean monthly water temperature.
WATEMP.EXE reads monthly air temperature files created using DATAIO.EXE and mean annual
wind speed file, calculates water temperature, and writes monthly water temperature to the
output. All input and output files are comma delimited.
WATEM.EXE is packaged as a windows application. It was executed using File/Run from the Windows program manager.
Example syntax: watemp.exe tempmean.txt comwind.txt watemp.txt,
where: watemp.exe = executable program; tempmean.txt = mean monthly air temperature;
comwind.txt = mean
annual wind velocity; watemp.txt = monthly water temperature.
An extract of WATEMP.TXT file is reported below:
Note: WATEMP.TXT was 214 Mb and took the PC 12 hours to create.
Changing the ASCII file TEMPMEAN.TXT to the format required by ARC/INFO
The QBASIC program AIRTEMP.BAS (above) was copied, renamed (WATEMP.BAS) and
used to change the output of WATEMP.EXE to the format required by the ARC command
ASCIIGRID.
Converting the output files of AIRTEMP.BAS to ARC/INFO GRIDS
After generating mean monthly water temperature files with WATEM.BAS, the 12 output files
were converted to UNIX format and then the ASCIIGRID command was used to convert ASCII
files to GRIDS. The AML airgrids.aml (step 1.10) was copied, and renamed (watergrids.aml)
to automate this procedure for the 12 months:
Table 8.9 shows the statistics on the 12 monthly water temperature values for Africa:
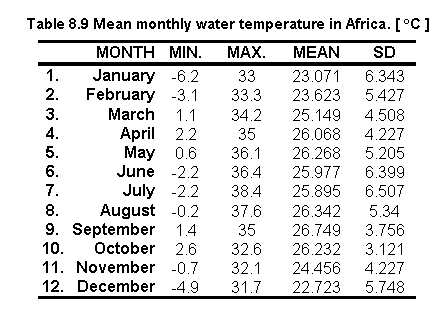
Reclassifying water temperature values to zero for temperatures below 0 oC
Because the water temperature model (watemp.exe) has been adequately tested only for
conditions where the water temperature exceeds 0 oC, the water temperature
below 0 oC should be replaced with mean monthly air temperature. However, by
isolating both the water temperature and the air temperature pixels < 0 oC
it was found that the corresponding air temperature values for these water temperatures
were also found to be < 0 oC. Moreover, by counting the number of pixels
< 0 oC for each image it was found that on average each image had 0.002
percent of pixels < 0 oC (highest number of pixels < 0 oC was
found in January with 0.007 percent).
The following sequence of GRID commands illustrates the procedure used to isolate and evaluate both the water and air temperature pixels which had values < 0 oC for the month of January:
The AML watemp_zero.aml automates the procedure for months 1, 2, 6, 7, 8, 11 and 12. The rest of the months ( 3, 4, 5, 9 and 10) did not have values < 0 oC so they remained the same.
Because it was not possible to replace the water temperature vales < 0 oC for air temperature it was decided to reclassify these values to zero:
The AML reclass_zwatmp.aml automates this procedure for all the moths (i.e. 1, 2, 6, 7, 8, 11 and 12).
Comparing mean monthly air and water temperature values
Results of the temperature ranges are presented in Table 8.10. Numbers in bold are
those months for which the water temperature values < 0oC had to be
substituted by a zero. The influence of annual wind velocity is also shown in this table
by comparing the air temperature with the water temperature values.
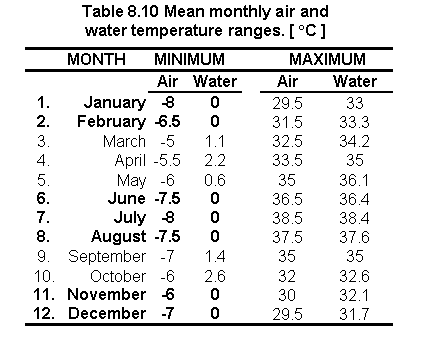
Fig. 8.17 Mean monthly distribution of water temperature values
The physical suitability of the considered species was evaluated by two programs called FISHSIM.EXE (i.e. Common carp and African catfish) and TILAPIA.EXE (i.e. Nile tilapia) which both require, for input, the mean monthly temperature file (WATEMP.TXT). These programs match the water temperature of each cell location with the fish requirement database (NT-PARAM, PARAMS.DB, TILAPIA.DB and SPECIES.DB).
The "NT-PARAMS.DB" file contains the simulation parameters for Nile tilapia:
The first line is the header indicating the internal names of the parameters. The next two lines are for the commercial and subsistence runs for Nile tilapia. The relevant commercial and subsistence output files are til-com.txt and til-sub.txt.
The "PARAMS.DB" file contains the simulation parameters for Common carp and African catfish:
The first line is a header indicating the internal names of the parameters. The next two lines are for the commercial and subsistence runs for Common carp, the same for African catfish. The relevant commercial and subsistence output files are carp-com.txt, carp-sub.txt, cat-com.txt and cat-sub.txt.
The "TILAPIA.DB" file has the bioenergetic parameters for Nile tilapia:
The "SPECIES.DB" file has the bioenergetic parameters for Common carp and African catfish:
From the run menu in Windows 95 the following commands were typed:
a) For Nile tilapia: tilapia.exe nt-params.db watemp.txt
b) For Common carp and African catfish: fishsim.exe params.db watemp.txt,
where: "watemp.txt" is the name of the relevant water temperature file.
Note: Data and programs are all contained within the same directory. SPECIES.DB and TILAPIA.DB are not typed in the dialog box of the run menu (section above), but they are used by their corresponding program during their execution.
At this stage, the programs automatically executed the scenarios indicated above, and created the corresponding output files:
Each output file contained the following data:
Note: It took the PC about 19 hours to run the FISHSIM.EXE programme and 12 for TILAPIA.EXE.
Changing the ASCII files (til-sub.txt, til-com.txt, cat-sub.txt, cat-com.txt, carp-sub.txt and carp-com.txt) to the format required by ARC/INFO
The QBASIC program AIRTEMP.BAS (step 1.9) was copied, renamed for each of the 6 txt files (carpcom.bas, carpsub.bas, catcom.bas, catsub.bas, tilcom.bas and tilsub.bas) and used to change the outputs of FISHSIM.EXE and TILAPIA.EXE to the format required by the ARC command ASCIIGRID.
Converting the output files of AIRTEMP.BAS to ARC/INFO GRIDS
The 12 output files were converted to UNIX format and then the ASCIIGRID command was used to convert ASCII files to GRIDS. The AML airgrids.aml (step 1.10) was copied, and renamed (fishgrids.aml) to automate this procedure:
Reclassifying into four suitability classes
The fish yield grids were reclassified into 4 equal-interval classes (quarters) assigning number 1 to the lowest range and 4 to the highest. Values assigned with a zero representing no crops retained the zero value. Example for small-scale fish yield of Common carp:
Grid: re_cpsub1 = slice (cpsub1, table, remap_psub1),
where: re_cpsub1 = reclassified grid; slice = GRID command to reclassify real numbers; cpsub1 = original grid; table = part of slice GRID command; re_cpsub1 = lookup table for reclassification.
This appendix briefly summarizes refinements that have been made to the fish growth simulation model used to assess inland aquaculture potential in Latin America (Kapetsky and Nath, 1997). Additional details regarding the original model can be found elsewhere (Nath, 1996). The refined model has been incorporated into POND Version 4.0, the decision support system developed at the Department of Bioresource Engineering, Oregon State University.
Model structure
The fish bioenergetics model used by Kapetsky and Nath (1997) considered the effects of fish size, food availability, photoperiod, and temperature on growth performance. This model was refined in the current study to address the effects of (i) high fish biomass, and (ii) feed type (moisture, protein and energy contents) and feeding levels on fish growth.
High biomass effects on fish growth in fed ponds
It is known that fish growth may often be reduced in fed ponds that have a high fish biomass (Hepher, 1988). The actual mechanisms by which these effects occur include deterioration of water quality and behavioral changes, neither of which can be adequately addressed without adding substantial complexity to the structure of the bioenergetics model. The alternate approach used in the current study involves the definition of a species-dependent parameter, the critical fish biomass (CFBfeed) which is the standing crop below which growth is not adversely affected in fed ponds. To account for high fish biomass (FB) effects, daily fish growth rate (g d-1) calculated in the bioenergetics model (Kapetsky and Nath, 1997) is multiplied with a biomass scaler (B; 0-1) given by:
Effects of feed type and feeding levels on fish performance
A fundamental assumption of the earlier version of the fish bioenergetics model (Nath, 1996; Kapetsky and Nath, 1997) was that the composition of fish and their diet is identical. This assumption has its roots in Ursin's (1967) growth model, which is the basis of our work in the area of fish bioenergetics. However, to adequately account for the effects of supplemental feeds of different quality (primarily moisture, protein and energy contents) on fish performance, it is necessary to remove the above assumption. Further, the bioenergetics model used by Kapetsky and Nath (1997) assumed that the digestibility coefficient is constant. In reality, this parameter varies with fish feeding levels (Meyer-Burgdorff, Osman and Gunther, 1989). In this section, we discuss changes made to the bioenergetics model (Kapetsky and Nath, 1997) to account for the effects of the above variables.
Moisture Content: In the present version of the bioenergetics model, the ratio of the dry matter content of fish (DMfish; g dry matter per g fish) to that of feed (DMfeed; g dry matter per g feed) is used to adjust the daily ration (R; g feed per day) for differences in moisture content between fish and the feed material supplied. The expression used is:
![]() (2)
(2)
where W = fish mass (g), fs = fraction of the diet that comprises artificial feed, and q = feed quality coefficient.
Protein content: According to Hepher (1988), dietary protein (dp expressed on a % dry matter basis) does not affect fish growth if it is above a critical level (Pcrit) that is species dependent. As dietary protein level reduces from Pcrit, growth drops off at an increasing rate. The following protein scaler (Ps; 0-1) used in the current study captures this effect:
![]() (3)
(3)
where p1 is a parameter that controls the rate at which Ps changes as dp decreases.
Energy content: A similar approach to the one for protein is used in the bioenergetics model to account for situations where dietary gross energy drops below a critical level (Ecrit; kcal g-1) that is again species dependent. The corresponding equation is as follows:
![]() (4)
(4)
where Es = energy scaler (0-1), and p2 is a parameter that controls the rate at which Es changes as the gross dietary energy de (kcal g-1) decreases.
It is difficult to evaluate growth response to diets that are sub-optimal in both protein and energy contents. For simplicity, we assume that when both protein and energy are below the respective critical levels required by the species, the scaler that is most limiting reduces the anabolic term in the bioenergetics model (see Kapetsky and Nath, 1997 for additional details regarding estimation of the anabolic term.).
Feeding levels and digestibility: It is well established that the digestibility (b) of food decreases as the amount consumed by fish increases (Hepher, 1988; Meyer-Burgdorff, Osman and Gunther, 1989). In the present bioenergetics model, we assume that digestibility decreases linearly with increasing levels of feeding (from maintenance to full satiation). The slope (e) of this relationship is estimated as follows (Figure 8.18):
![]() (5)
(5)
where bmax = maximum digestibility coefficient (assumed to occur at a maintenance ration), bmin = minimum digestibility coefficient (at satiation), and fmaint = feeding level parameter in the bioenergetics model (Kapetsky and Nath, 1997) corresponding to a maintenance ration. Once the slope e from Equation 5 is estimated, the actual digestibility coefficient is obtained as follows:
![]() (6)
(6)
where f = feeding level parameter (0-1) for the actual feeding rate.
Model calibration and validation
In order to use the revised bioenergetics model (i.e., including equations 2-6), it was necessary to estimate six new parameters (i.e., Pcrit, Ecrit, p1, p2, bmax and bmin), in addition to the previous one in the fish bioenergetics model (Kapetsky and Nath, 1997). Among the six parameters, Pcrit and Ecrit were adapted from bioenergetic studies with different species as synthesized by Hepher (1988). The other four new parameters were added to an automatic model calibrator (Nath, 1996), which estimates appropriate values by the use of an adaptive, non-linear search algorithm in conjunction with experimental data from pond trials.
A listing of the 15 parameters estimated for the three fish species in this study is provided in Table 8.11. Data sources and model performance for each of the species are discussed below.
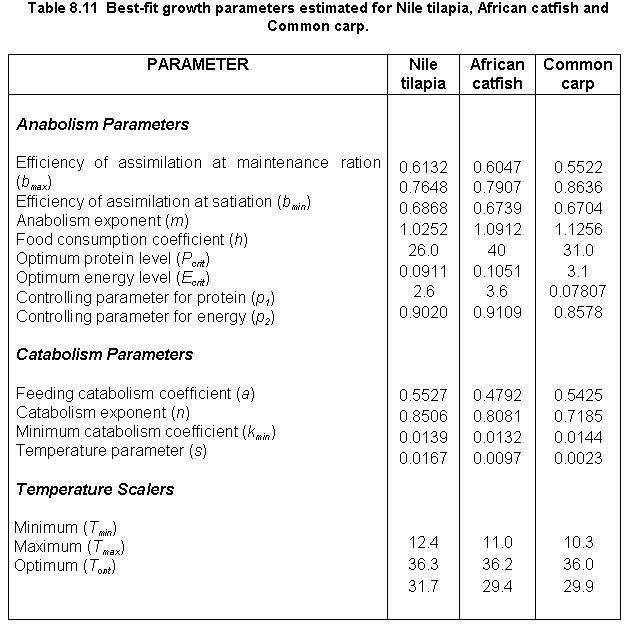
Nile tilapia
A large number of datasets appropriate for model calibration and validation was collected for this species. Consequently, calibration of the energetics model for this species was accomplished across several sites and different production systems, ranging from a small-scale fertilized system in Ghana to intensely fed ponds in Thailand (Table 8.12). Parameters estimated for this species (Table 8.11) resulted in fairly good predictions of Nile tilapia growth at all the sites (Table 8.12). Model predictions are compared to observed data for two of the sites (Rwanda and Honduras) in Figure 8.19.
Model fits were not as accurate as those obtained with the previous version of the energetics model (Nath, 1996), presumably because the current study focused on generating a set of Nile tilapia growth parameters that would result in reasonable predictions at different geographical locations. This is in contrast to our previous study (Kapetsky and Nath, 1997) where model calibration for Nile tilapia was confined to only one site (El Carao, Honduras).
African catfish
Calibration for this species was accomplished by the use of growth data from sites in South Africa and the Central African Republic (CAR) (Table 8.12). Although growth predictions for both sites are quite good (Figure 8.20), there appears to be a slight tendency towards over-prediction of final fish weights (Table 8.12). This is particularly noticeable for the CAR simulation run, and appears to be due, in part, to a sharp reduction in observed fish weights from the expected growth profile (De Kimpe and Micha, 1974). Such departures are not uncommon in pond experiments and have been attributed to biases in sampling methods (Hopkins, 1992). It is difficult to account for such biases during calibration of the energetics model used herein without extensive knowledge and familiarity with actual experimental protocols.
Independent validation was not conducted for African catfish because only two datasets that were complete in terms of fish growth, temperature and feeding/fertilization data could be identified. These were both used for model calibration in order to generate a set of parameters that would reflect growth performance of this species at geographically different locations.
Common carp
The energetics model for this species was calibrated with data from Poland primarily because only one complete growth dataset for this species could be identified for African conditions. This dataset was reserved for model validation. Model performance was excellent for the two sites in Poland and South Africa (Figure 8.21; Table 8.12). It is expected that good predictions would be obtained at other locations as well because the above two locations are very diverse particularly in terms of ambient water temperatures.
Summary
A previously developed energetics model (Nath, 1996) was refined to include the effects of high fish biomass, feed types and feeding levels on growth performance of pond fish. Calibration and validation for three target fish species (Nile tilapia, African catfish and Common carp) was also successfully accomplished. In contrast to calibration procedures used previously that involved fitting the model to growth data for one location, we estimated parameters for the refined model across different sites. This approach is expected to result in better projections of fish growth for all three fish species, under different geographical conditions in the African continent.

Two models, based on multiple criteria analysis, were developed for the fish farming potential assessment:
a. Small-scale fish farming
b. Commercial fish farming
Inputs for the small-scale fish farming model were:
![]() Water requirement
Water requirement
![]() Soil and terrain suitability for ponds
Soil and terrain suitability for ponds
![]() Inputs
Inputs
![]() Farm-gate sales for aquaculture
Farm-gate sales for aquaculture
Inputs for the commercial fish farming model were:
![]() Water requirement
Water requirement
![]() Soil and terrain suitability for ponds
Soil and terrain suitability for ponds
![]() Inputs
Inputs
![]() Farm-gate sales for aquaculture
Farm-gate sales for aquaculture
![]() Urban market size and proximity
Urban market size and proximity
Standardisation and thresholds
Input criteria were evaluated in terms of suitability by assigning scores to the pixel values. A standard classification was applied to the layers used by the two models:
4 = Very suitable
3 = Suitable
2 = Moderately suitable
1 = Unsuitable
Small-scale model fish farming model
The formula was the following:
small = (waterlos_sl * 0.30 ) + ( soilslope_sl * 0.13 ) + ( byprod_sl * 0.04 ) + (farmgate_sl * 0.07 ) + ( mkgrd_sl * 0.46 )
Commercial model fish farming model
This grid was produced using the following formula:
comer = (waterlos_sl * 0.56 ) + ( soilslope_sl * 0.26 ) + ( byprod_sl * 0.12 ) + (farmgate_sl * 0.06 )
where: waterlos_sl = water requirement; soilslope_sl = soil and terrain suitability for
ponds; byprod_sl = potential for inputs; farmgate_sl = farm-gate sales; mktgrd_sl = urban
market size and proximity.
Integrating the models
The small-scale fish farming grid (small) was combined with the fish yield outputs for Nile tilapia (tilcom1), African catfish (catcom1), and Common carp (cpcom1) and the commercial grid (comer) was combined with the fish yield outputs for Nile tilapia (tilsub1), African catfish (catsub1), and Common carp (cpsub1).
The following procedures were used for grid combinations:
a) The grids small and comer were each multiplied by a factor of ten:
b) The grids were added to the fish yields:
The VALUE of the output grids created was a two digit integer in which the first digit indicated the class of the commercial model and the second the class of yield in terms of crops/y for each fish species.
c) The grids were re-classified to exclude those areas of no coincidence. Example for small-scale farming of tilapia.
Grid: rm_tilsub1 = reclass(md_tilsub1, remap_tilsub1),
where: rem_tilsub1 = reclassified image and remap_tilsub1 = table which assign the
values to re-classify).
Small-scale fish farming
a) Grid: smallcoi = (md_catsub1) + (md_cpsub1 * 10,000) + (md_tilsub1 * 100)
The VALUE of the output grid created was a six digit integer in which the first two digits indicate the class of grid md_catsub1; the second two digits indicate the class of grid md_cpsub1, and the last two digits indicate the class for md_tilsub1.
b) The grids were re-classified to exclude those areas of no coincidence.
Grid: smallcoi_sl = reclass(smallcoi, remap_smallcoi)
Commercial farming
Same procedure as for small-scale farming was followed:
a) Grid: comercoi = (md_catcom1) + (md_cpcom1 * 10,000) + (md_tilcom1 * 100)
b) Grid: comercoi_sl = reclass(comercoi, remap_comercoi)
Verification by existing fish farm locations
a) Creating the point features
Lat/Long locations of existing fish farms for Zimbabwe and Kenya were added to a coverage.
The Arc module GENERATE was used to add features to a coverage and the coordinates of each
feature were entered from a file:
where: generate = Arc module to create coverages; z1farms = output cover for farm number one; z1farms.txt = file containing Lat/Long coordinates of farm number one; points = feature cover type.
The same procedure was applied to Kenya, and those coverage's are located in the directory: /pepe57/verify/kenyaf/.
b) Verification
To obtain the suitability score from each one of the farm sites, the point cover created
with the generate command (above) was plotted over each of the resulting suitability
grids. The suitability score was obtained from each grid cell site where a farm was
located. Example of verification procedure for the water requirement suitability grid:
where: waterlos_sl = water requirement grid; z1farms = coverage containing Lat/Long location for farm number one; cellvalue = Grid command to query the grid value.
Verification by number of farms at a county-level
a) Digitizing
Arua counties were digitized from a paper map (Department of Lands and Surveys, 1986). A
detailed description of the methodology (10 pages) is provided in the document called digit.doc
located in the directory /sun2disk5/faogis7/pepe57/digit/ of SUN 2.
b) Verification
The grid file that contains all counties in Arua district was converted into an equal area
projection, and then statistical data was extracted following the same procedure described
in Appendix 8.11 for grid files.
Country grids and verification
Grids for Zimbabwe, Kenya and Malawi were extracted (i.e. by means of reclassifications) from the grid AFBOUNDARY which contains all the country areas. Each of the country grids (i.e containing only a value of one) were then multiplied to each one of the water temperature grids and then, the statistical water temperature data were extracted:
Grid: watmp1 * zimbabwe
INFO> SEL ZWATMP1.STA
where: watmp1 = water temperature grid for January; zimbabwe = zimbabwe grid (i.e. grid with a value of one); SEL ZWATMP1.STA = INFO syntax to extract statistical data; ZWATMP1.STA = water temperature grid for Zimbabwe for January.
The concordance coefficient W is based upon the following hypothesis:
HO: The m sets of rankings are not associated;
H1: The m sets of rankings are associated and are derived using the following formula:
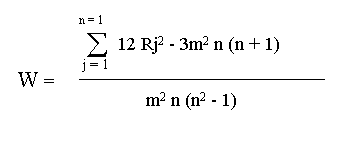
When the observed sets of rankings were in close agreement , W was large (close to one); conversely, when the agreement was poor W was close to zero. Therefore large values of W rejected HO. Furthermore, it was possible to compute: X2 = m ( n - 1) W, and compare it with the value of chi: x2 = (n - 1). If the X2 was larger than chi, rankings were associated and therefore there was an agreement.
High or significant values of W were interpreted as meaning that the decision-makers were essentially applying the same standard in ranking the factors under study. However, a significant value of W did not necessarily mean that the orderings observed were "correct". In fact, they may all be incorrect with respect to some external criterion (Siegel, 1965). It is possible that a variety of decision-makers can agree in ordering objects because they all employ the "wrong" criterion. In this case a high or significant W would simply show that all more or less agree in their use of a "wrong" criterion. To solve this problem Kendall (1984a) suggests that the best estimate of the "true" ranking when W is significant is provided by the order of the various sums of ranks, Rj.
A programme was created in MINITAB for Windows to automate the use of the formula above. However, a less simplified version of this formula was used because it was better suited for programming in MINITAB.
Simplified version of the Kendall coefficient of concordance formula:
If n items are ranked by m judges, and Xij denotes the rank number given to the ith item by the jth judge, then Kendall's concordance is represented by:
n
Sw =S ( Xi -
1/2 m (n + 1) )2,
i+1
where: Xi = SXij is the total of the m ranks given to the ith item and m(n+1) is the mean of the Xi. The maximum of value of Sw is m2 (n3 - n) / 12, representing perfect agreement between judges.
W = 12 Sw / m2 (n3 - n)
For details about this formula see: Greenwood and Hartley (1962).
MINITAB programme that automates the use of the Kendall Coefficient of concordance formula:
Converting grids to an equal-area projection
The result grids were converted to an equal-area projection (Flat Polar Quartic) to calculate the areas covered by each class in each country of Africa.
The grid AFBOUNDARY, which contains the country boundaries (i.e. areas) was converted to an equal area projection and was named COUNTRYGRD_PQ. The VALUE of the later grid is the country code. This grid was overlaid on the various themes in order to produce the statistics by country.
Arc: project grid afbounday countrgrd_pq
To convert grids to Flat Polar Quartic projection the parameters specified in the file llpolq.prj illustrated below were used:
- INPUT
- PROJECTION GEOGRAPHIC
- UNITS DD
- PARAMETERS
- OUTPUT
- PROJECTION FLAT POLAR QUARTIC
- UNITS METERS
- PARAMETERS
- 00 00 00
- end
Combining grids
a) Combining COUNTRYGRD_PQ with the result grids.
The following is an example for the water loss grid:
Grid: cnwaterlos = combine(countrygrd_pq, waterlos_pq),
where: cnwaterlos = combined grid; combine = GRID command; countrygrd_pq = African country
boundaries in flat polar quartic; waterlos_pq = water loss grid in flat polar quartic.
Adding the country names
a) The country name item (CNTRY_NAME) was added to the new grid:
Arc: additem cnwaterlos.vat waterlos.vat cntry_name 40 40 C
Where: 40 40 C indicate the number of words to be used in the country name item.
b) Adding the country names:
To add the country names according to the country values in COUNTRYGRD_PQ in the country item (CNTRY_ITEM) the AML cnnames.aml was used:
Creating the text file
Manipulating text files in EXCEL
a) The text files were imported into EXCEL
b) To calculate the areas of each class occurring in the countries, the number of cells (COUNT) was multiplied by the square of the cell size in kilometers (5.930642 * 5.930642).
c) Percentage areas were calculated from each class for each country.
d) Results were plotted using histograms.
File names
For clarity, Grid and file names were coded in such a way that names portray the essential information about the contents of the grids.
Models and statistics
Grids
Digits
Content Meaning
Digits 1 - 3
md_ Model
rm_
Reclassified grid
Digits 4 - 6
til
Tilapia
cat
Catfish
cp
Carp
Digits 7 - 9
sub
Small-scale fish farming
com
Commercial fish farming
Digits 10
1
Crop/year
The procedures used to create map compositions of single or annual grid files are illustrated below using the water requirement grid as an example:
Standardising the grid to a common colour range
For purposes of analysis and/or illustration, the single or annual grid files had to be reclassified to a common colour range. The following example is illustrated for mean annual wind velocity:
cwater = reclass (water, water.rem)
where: cwater = reclassified grid file; reclass = GRID function to reclassify (or change) integer values of the input cells using a remap table on a cell-by-cell basis within the analysis window; water = original water requirement grid; water.rem = remap table.
The remap table water.rem is shown below:
- >0 : 1
- -2,000 -1 : 2
- -2,000 -3500 : 3
- > -3500 : 4
Preparing the grid file for plotting
Preparing the keyshade.
To create the colour range legends in accordance to the remap table water.rem above the text file water.txt was created:
- .1
- >0
- .2
- -2,000 -1
- .3
- -2,000 -3500
- .4
- > -3500 mm
AML to plot the grid
The AML water.aml illustrated below was used in ARCPLOT to generate a map
composition water_mc for the water requirement grid:
Note: The AML water.aml was used for all criteria (e.g. soils, inputs, farm-gate sales, mean annual wind velocity,) and only some elements (i.e. reclassification AML's, remap tables and keyshades) needed to be changed for each criteria according to their range of values.
In cases where the input grid values were real the " SLICE" GRID function was used (e.g. net annual water requirement) and in cases where the input values were integers the "RECLASS" GRID function was used (e.g. mean annual wind velocity).
The procedures used to create map compositions of monthly grids for precipitation, potential evapotranspiration, water requirement, air temperature, water temperature, are illustrated below using the precipitation grids as an example:
Standardising grids to a common colour range
Most of the grids created had a large number of values, so it was necessary to reduce this number to facilitate interpretation and analyses. The following example illustrates how one of the GRID files for precipitation was reclassified:
crain1 = reclass (rain1, rain1.rem)
where: crain1= reclassified precipitation grid for the month of January; reclass = grid function to reclassify the values of the input cells by specified ranges; rain1.rem = remap table.
A GRID AML rerain automates this procedure for all monthly precipitation grids by repeating the command above for each one of the GRID files.
The remap table rain.rem is shown below:
Preparing grids for plotting
a) Generating a stack from the 12 monthly precipitation GRID files.
To group (or stack) the 12 monthly grids, the GRID command MAKESTACK was used:
Grid: makestack rainstack list crain1 crain2 crain3 crain4 crain5 crain6 crain7 crain8 crain9 crain10 crain11, crain12 where: makestack = GRID command; rainstack = name of the output stack; list = keyword indicating that the grids to be used to generate the stack will follow on the input command line; crain1 to crain12 = names of input grids that make up the stack.
b) Preparing the keyshade
To create the colour range legends in accordance to the remap table rerain above
the text file rain.txt was created. Note that the constraints were assigned a value
of 27.
AML to plot the monthly grid files
The AML rain.aml illustrated below was used in ARCPLOT to generate a single map
composition rain_mc for the 12 mean monthly precipitation grid files:
Note: The AML rain.aml was used for all criteria (i.e. precipitation, potential evapotranspiration, water requirement, air temperature and water temperature,), and only some elements (i.e. reclassification AML's, remap tables, stacks and keyshades) needed to be changed for each criteria according to their range of values.
In cases where the input grid values were real the " SLICE" GRID function was used (i.e. air temperature, water temperature and water requirement) and in cases where the input values were integers the "RECLASS" GRID function was used (i.e. precipitation and potential evapotranspiration).
Shadesets
Three palettes (shadesets) were created to represent the result grids. The first shadeset named pepe4.shd (Table 8.13) was used to represent the colour range for those grids used in the farming system models and the fish growth model which contained 5 or 6 values. The shadeset pepe44.shd was used for the combined models and coincidence grids. The third shadeset, grey5.shd was used to illustrate all maps in the appendix. Shadesets were created using the SHADEEDIT command in ARCPLOT.

Postscript files
All grid files which are presented in this study were converted to eps (i.e. postscript format). The following example illustrates how the water requirement grid was converted into and eps file:
where: setenv CANVASCOLOR WHITE = sets the background color to white; &r water.aml = automates the map composition of the grid; display 1040 = command used to save the map composition; water.gra = graphics file; plot water_mc box 0.5 0.5 7.7 10.7 = map composition which was reduced to suit the size required for publication; postscript = ARC command used to create the postscript file.